ASTER-REP is the first transposable elements (TEs) database of full-length sequences belonging to Asteraceae species. One feature that differentiates this database from the others is using full-length sequences instead of prototypic sequences. ASTER-REP significantly improves transposon annotation for the species already present in the database and other Asteraceae genomes; one of our goals is to continuously update ASTER-REP by adding TEs identified in species whose genome has been fully sequenced.
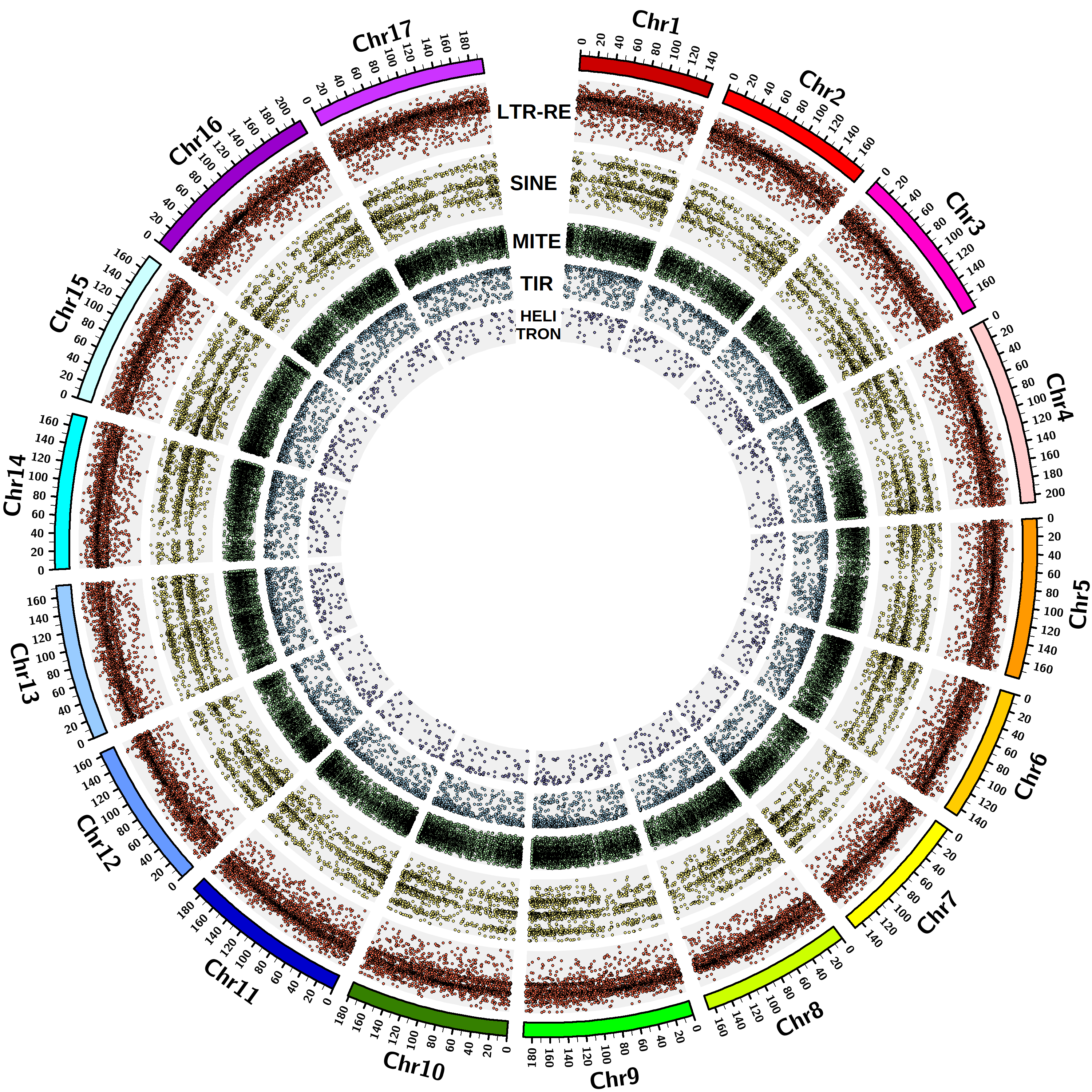
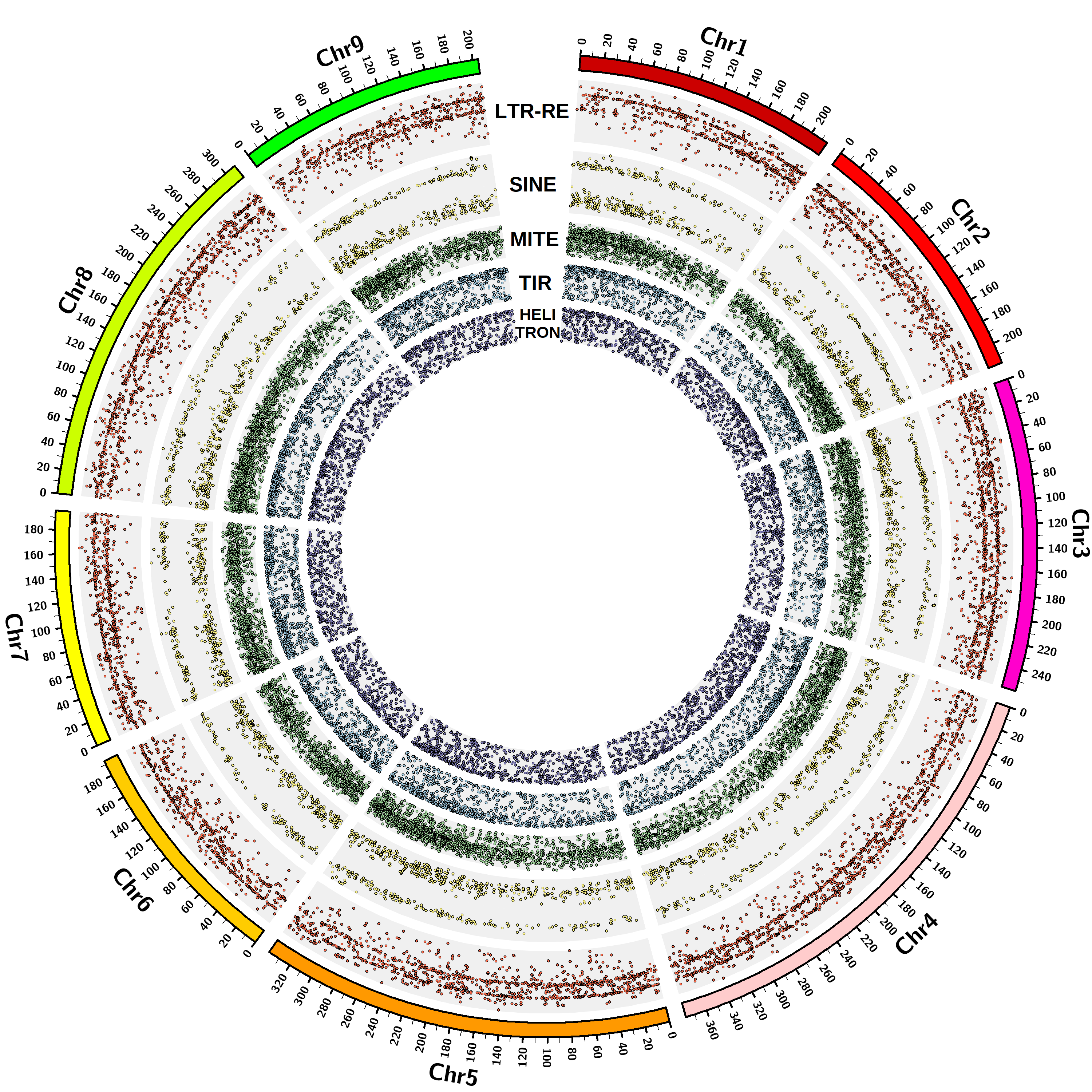
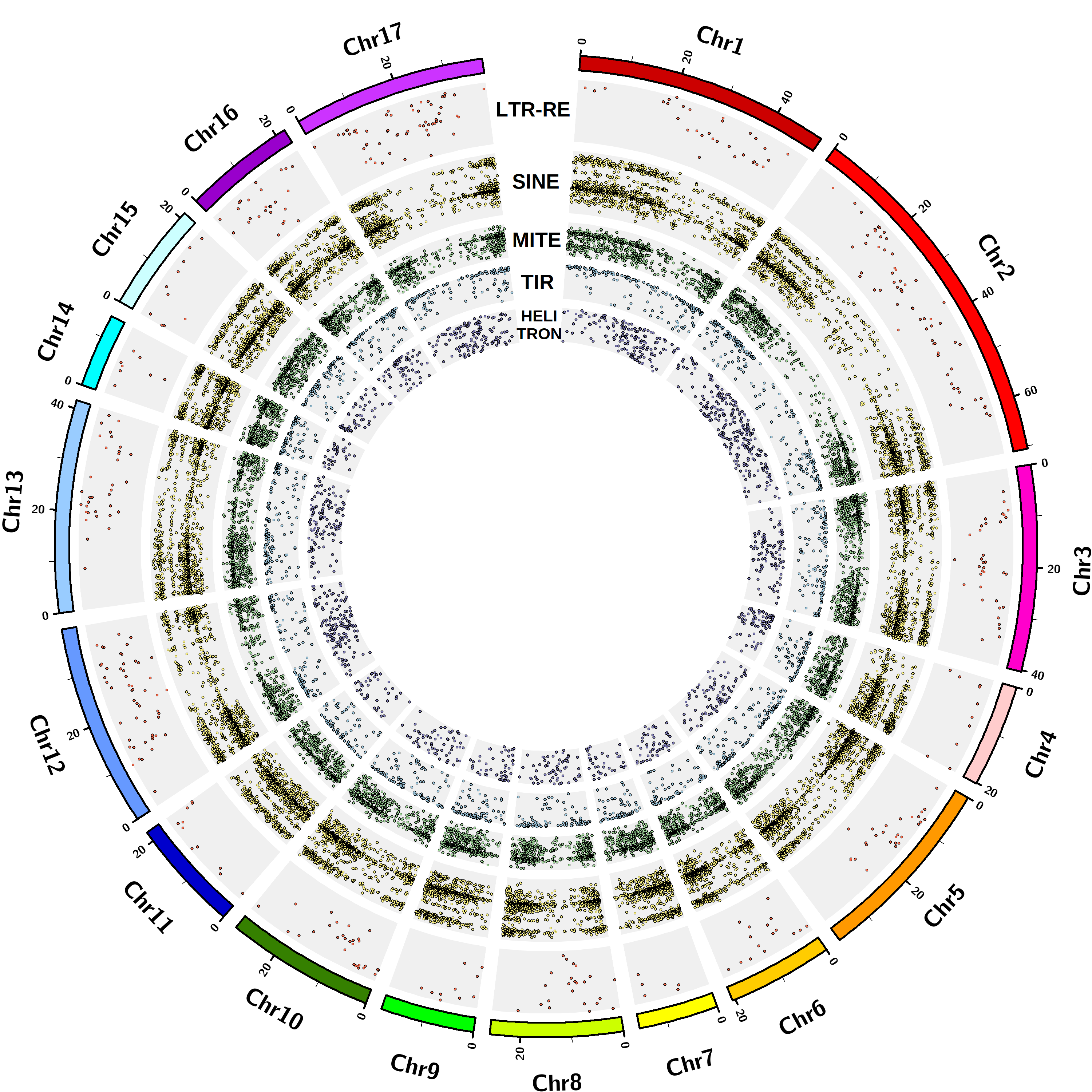
The TE orders identified to date in the Asteraceae genomes are long terminal repeat (LTR) and small interspersed nuclear element (SINE) for Class I TEs, terminal inverted repeat (TIR), miniature inverted-repeat transposable element (MITE) and Helitron for Class II TEs, according to the most recently proposed TE classification. The elements were identified using software based on structural features recognition [Ventimiglia et al …]. A total of 328,696 full-length TEs are currently included in the database.
ASTER-REP is set up on a Linux-Apache-MySQL-PHP (LAMP) system. JavaScript libraries, including jQuery 3.6.0, Bootstrap 5.1.3, Datatables 1.11.3 and some additional plugins to perform dynamic web services were used. Users can obtain the desired data with automatic search options by selecting checkboxes for desired Species, whereas TE Class, Order, Superfamily and Lineage are selectable from drop-down menus. The search results, consisting of a multi-FASTA file and a GFF file relative to all the selected elements, can be downloaded locally using the temporary links provided via email.
The Asteraceae species presently included in the database are: Helianthus annuus, Lactuca sativa, Cynara cardunculus var. scolymus, Artemisia annua, Carthamus tinctorius, Chrysanthemum seticuspe. The corresponding whole genome assemblies, on which the analyses for TE discovery were conducted, are available on the National Center for Biotechnology Information (NCBI).
| Species | No. of chromosomes | Assembly size (Gb) | GenBank assembly accession | Assembly level | No. of TEs identified |
|---|---|---|---|---|---|
| Helianthus annuus | 2n=2x=34 | 3.01 | GCA_002127325.2 | Chromosome | 125,662 |
| Lactuca sativa | 2n=2x=18 | 2.39 | GCA_002870075.2 | Chromosome | 32,303 |
| Cynara cardunculus var. scolymus | 2n=2x=34 | 0.72 | GCA_001531365.1 | Chromosome | 30,556 |
| Artemisia annua | 2n=2x=18 | 1.79 | GCA_003112345.1 | Scaffold | 54,711 |
| Carthamus tinctorius | 2n=2x=24 | 0.66 | GCA_001633085.1 | Scaffold | 31,217 |
| Chrysanthemum seticuspe | 2n=2x=18 | 2.72 | GCA_004359105.1 | Scaffold | 54,247 |